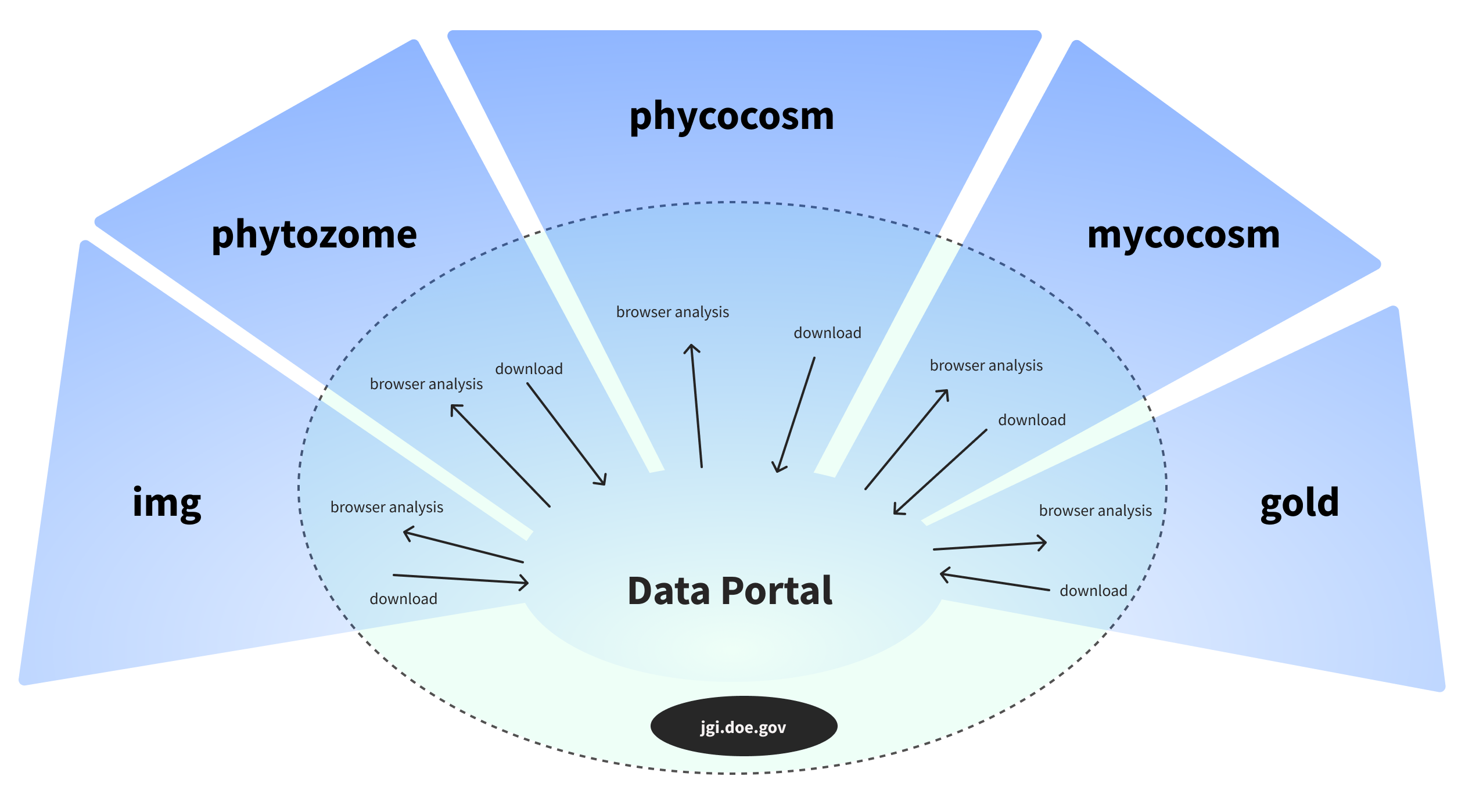
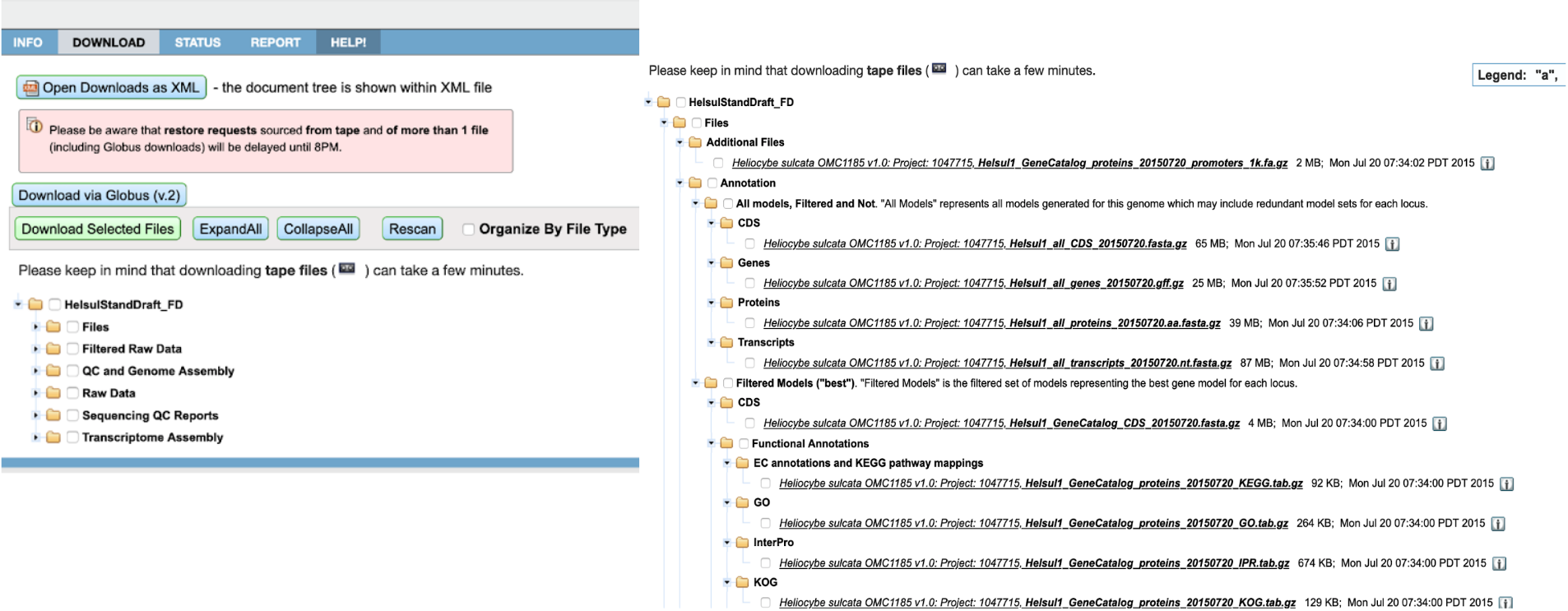
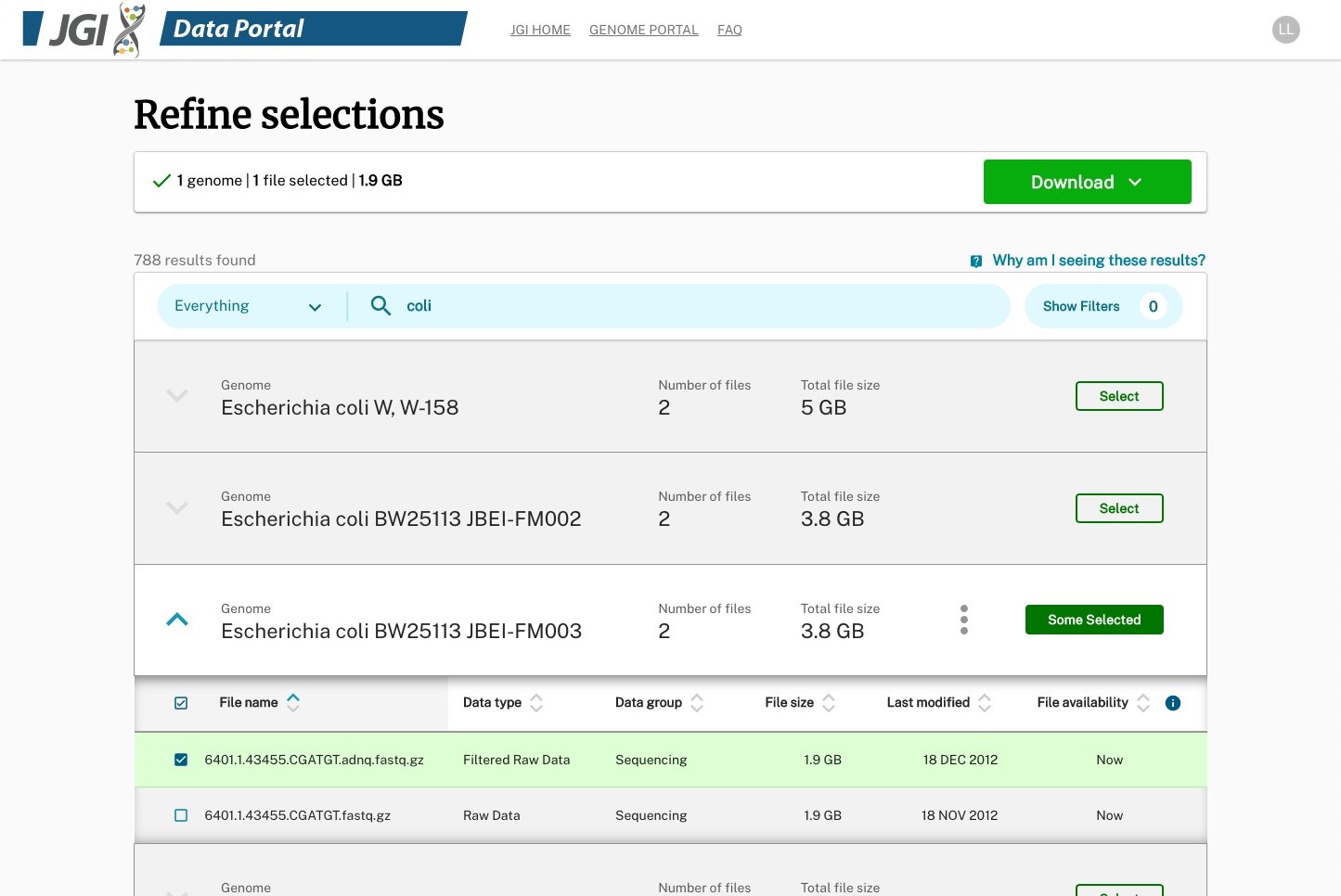
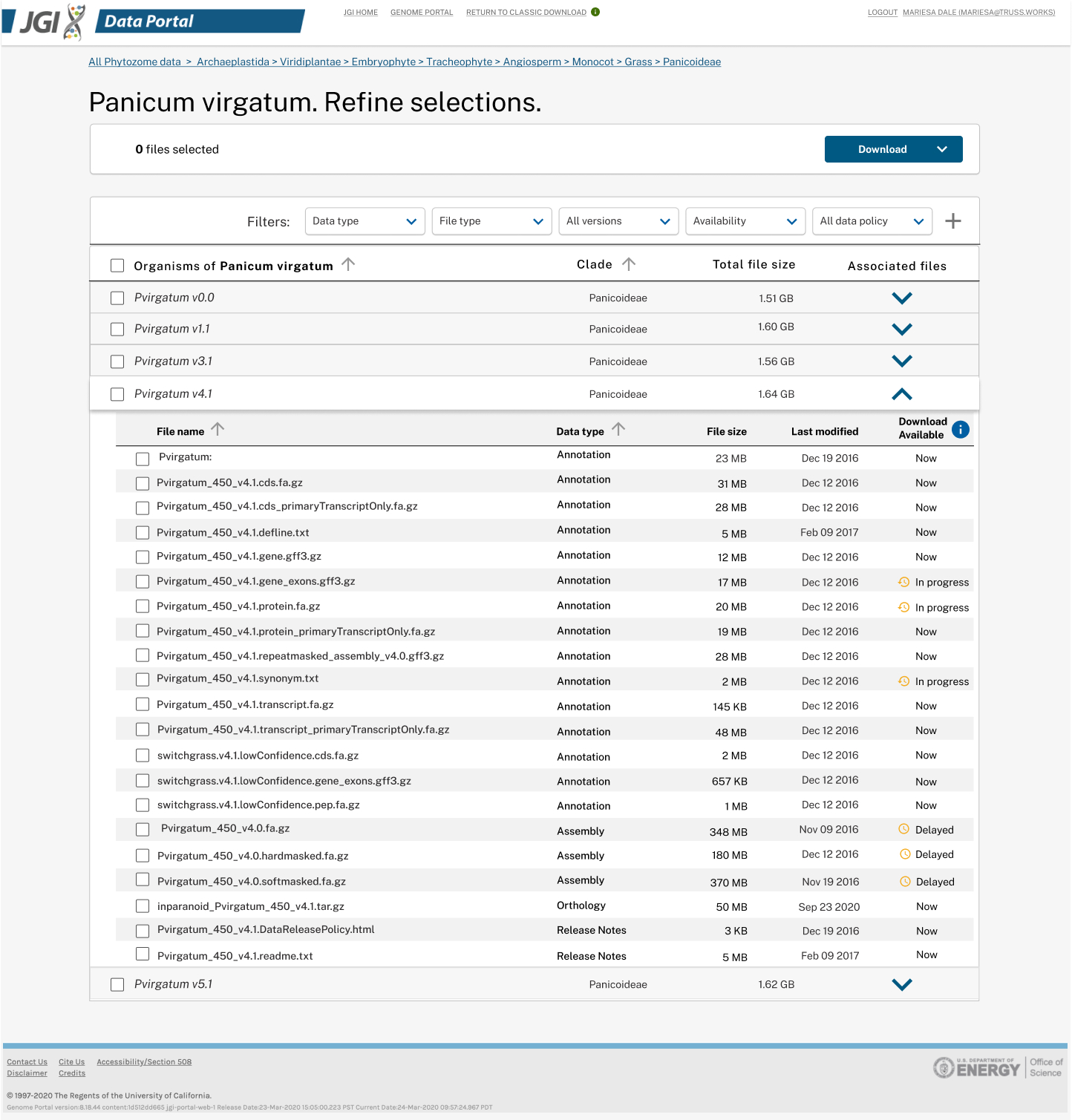
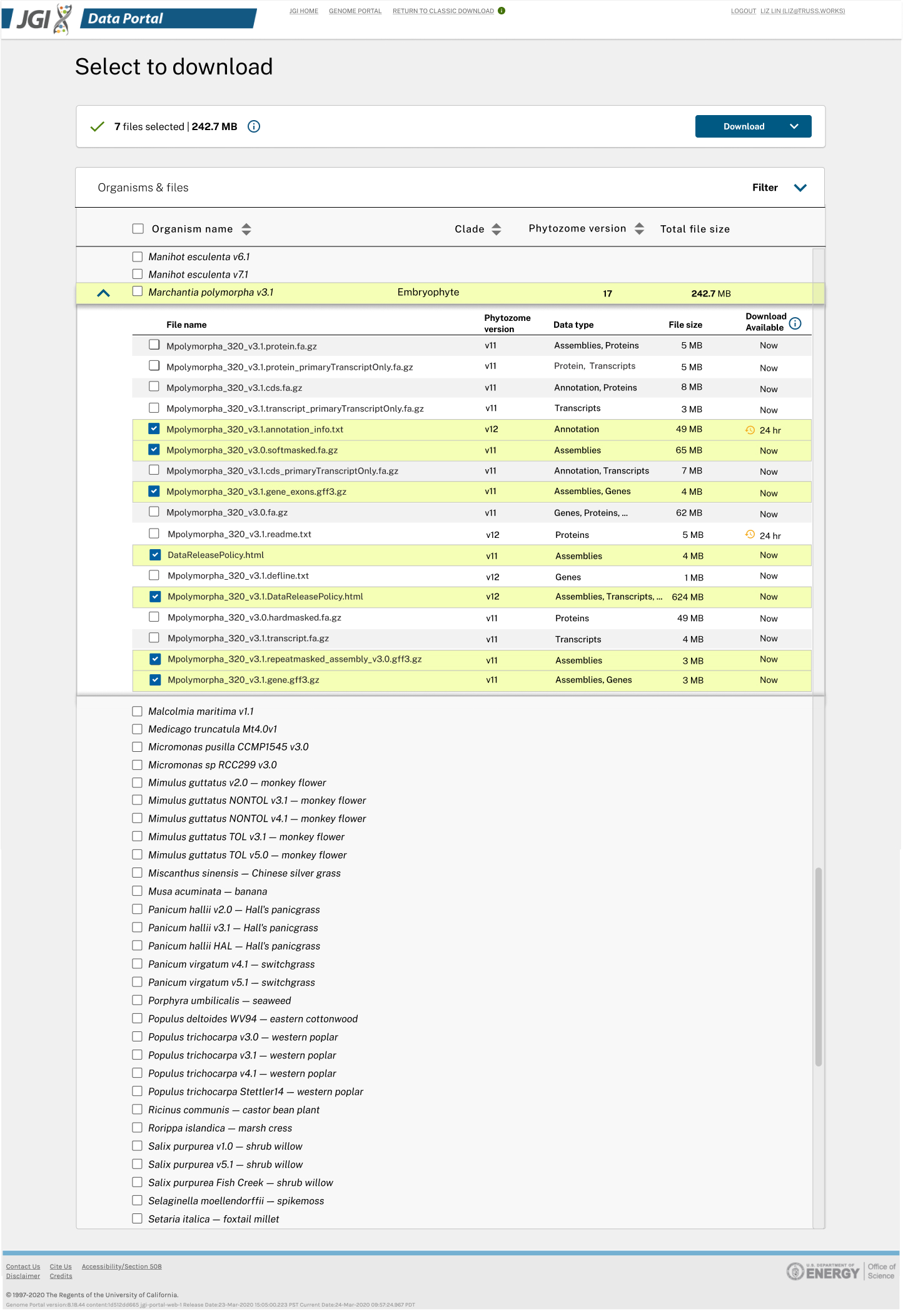
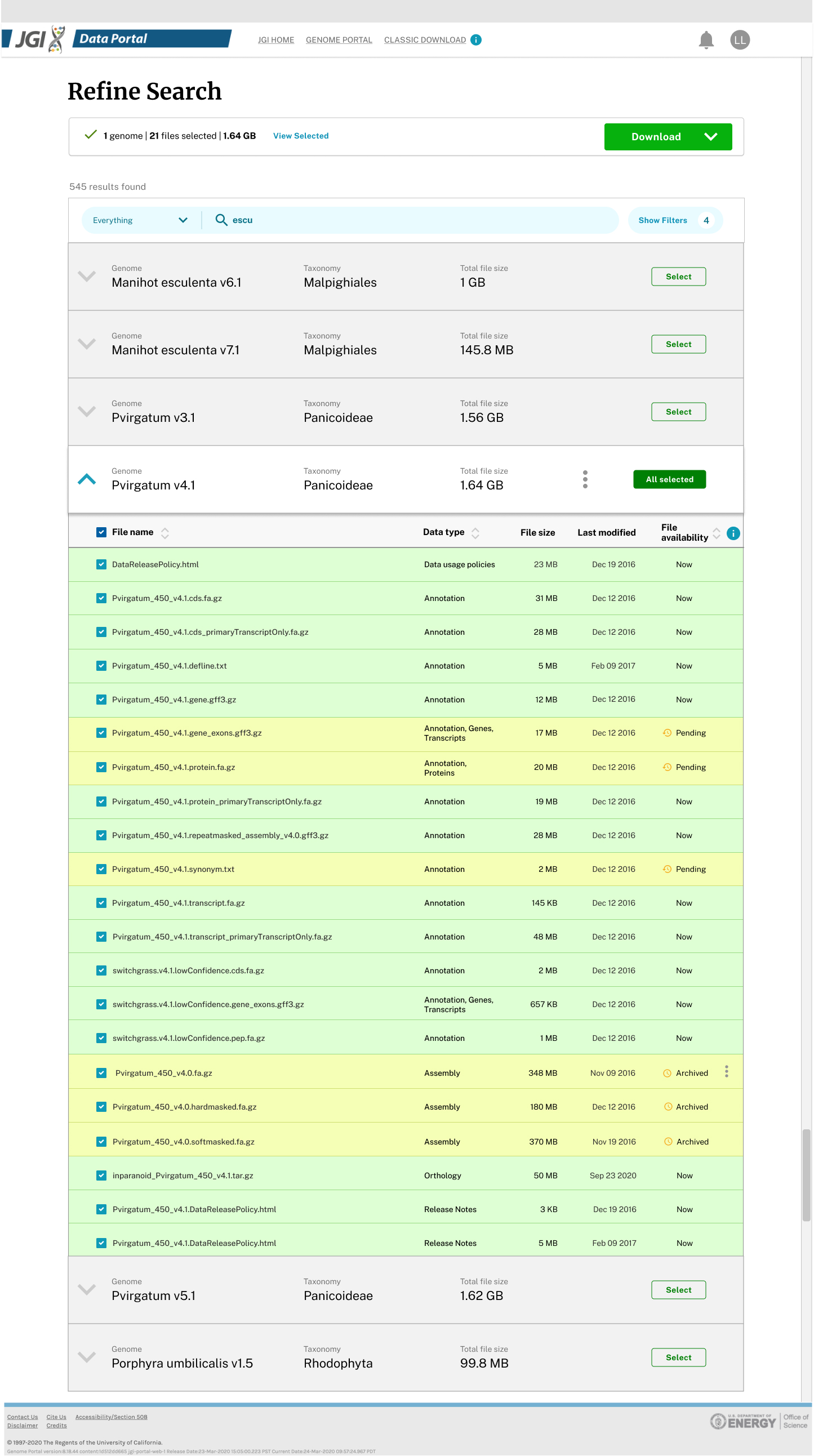
Started with single organism download, we created robust "filter and refine" process prior to downloading. Displaying all the available plant genomes in a tree made sense at the beginning.
With CI/CD and feature flags, we were able to test-in-production, conducting usability tests early and frequently. After multiple iterations, we became familiar with the underlying metadata and their strengths & weaknesses. We started to make suggestions, request updates, and share feedback with different kingdom teams.
We also realized that the Phylogenetic tree design would not scale to 1,000 fungal or 15,000 microbial datasets. The findings and insights from usability tests also supported us to switch from the tree view to the list view, for it reduced the number of steps scientists needed to view or download.